DSS: Redefining Biotechnology & Life Science in India
- About Us
- Products & Services
PRODUCTS & SERVICES
- Applications & Specialities
All Applications & Specialities
- Brands
- Contact Us
-

-
 0
0
- ☰
- About Us
- Products & Services
-
Kits Reagents & Consumables
- Cytogenetics
- Dyes
- Fluorescence In Situ Hybridization (FISH)
- High-Performance Liquid Chromatography (HPLC)
- Histology
- Immuno Histo Chemistry (IHC)
- IVF Consumables
- Molecular Pathology & Diagnostics
- Multiplex Ligation-Dependent Probe Amplification (MLPA)
- Nucleic Acid Extraction
- PharmDx
- Real Time PCR
- Special Stains
- Instruments
- Software
- Accessories
- Advanced Material
- Therapies
-
Kits Reagents & Consumables
- Applications & Specialities
- Brands
- Brand - Life Sciences
- 3i
- ABBERIOR INSTRUMENTS
- Abbott Molecular
- ADS Biotec
- APPLIED SPECTRAL IMAGING
- BioAir Tecnilabo
- DAKO (AGILENT)
- Eden Tech
- Elveflow
- ENTROGEN
- EUROCLONE
- EVIDENT
- Genea
- Hamamatsu Photonics
- Invivoscribe
- MASTER DIAGNOSTICA
- MBF BIOSCIENCE
- MBST
- Medical Tek Co. Ltd
- MILESTONE MED SRL
- Molecular Machines & Industries
- MRC HOLLAND
- NeoDx
- Onward Assist
- Profound
- SCIENTIFICA
- SpaceGen
- Seqlo
- µCyte
- Brand - Industrial
- Brand - Life Sciences
- News & Events
- Career
- Contact Us
- Testimonial
- Blogs
- R&D
- CSR
- Press Release

Beyond the Microscope: Detecting Genomic Changes with D024 KaryoProfiler
BY Ms. Megha Dhumal 17th February 2026
In the Quiet Hours of the Lab
In the quiet hours of a cytogenetics laboratory, cells are busy at work while no one is watching. They divide, adapt—and sometimes, silently, they change. By the time scientists return, metaphase spreads are ready—but did the genome remain stable overnight?

For decades, cytogeneticists tried to answer this question by peering through microscopes. Classical karyotyping laid the foundation for understanding genetic disorders, cancer, and chromosomal biology by revealing large structural chromosomal abnormalities. Yet as research moved into long-term cultures, stem cells, and high-throughput models, subtle but biologically significant chromosomal changes increasingly escaped detection.
From Karyotyping to Genome-Wide Insight: A Historical Journey
The First Milestone: Counting Chromosomes
In 1956, Joe Hin Tjio and Albert Levan corrected decades of misconception by establishing that humans have 46 chromosomes. This breakthrough immediately linked chromosomal abnormalities to disease, enabling the identification of trisomies, monosomies, and large structural defects. Early karyotyping could only detect large alterations (≈5–10 Mb), but it was a revolutionary start.
The Rise of Karyotyping
The introduction of G-banding in the 1960s allowed cytogeneticists to reliably distinguish individual chromosomes. Translocations, inversions, deletions, and duplications became visible, cementing karyotyping as a cornerstone of diagnostics for congenital disorders, such as Down syndrome, and hematologic malignancies. While this method opened new possibilities, it relied on dividing cells and lacked sensitivity for smaller copy number changes or low-level mosaicism.
FISH: Turning on the Spotlight
Fluorescence in situ hybridization (FISH), introduced in the 1980s, brought the genome into sharper focus. By tagging specific DNA sequences with fluorescent probes, scientists could visualize chromosomal changes—even in non-dividing cells. FISH confirmed suspected abnormalities, detected microdeletions, duplications, translocations, and gene amplifications, and highlighted clinically relevant targets, from rare syndromes like 22q11.2 deletions to cancer markers such as HER2 and ALK.
However, FISH only illuminates the regions that are specifically probed. To gain a more comprehensive view of the genome, researchers turned to genome-wide approaches, including MLPA, microarrays, and NGS-based copy number profiling. These methods enabled detection of subtle gains, losses, and low-level mosaicism across the entire genome—providing a broader perspective that targeted methods alone could not.
CGH and Microarrays: Seeing the Genome as a Whole
Comparative genomic hybridization (CGH) in the early 1990s enabled detection of large chromosomal gains and losses, though early methods were limited to metaphase chromosomes and low resolution. DNA microarrays, including aCGH and SNP arrays, dramatically increased resolution to approximately 50–200 kb, depending on platform and probe density, revealing submicroscopic deletions and duplications. While powerful, these methods require specialized expertise and can be costly, creating the need for fast, reliable, genome-wide solutions. MRC Holland addressed this challenge with the NXtec D024 KaryoProfiler, providing sensitive, comprehensive copy number profiling across the genome.
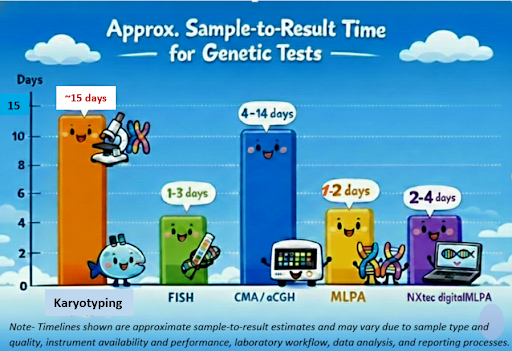
The Modern Challenge: Genomic Stability
Even in carefully maintained cultures, the genome is never static. Over time, subtle chromosomal changes can quietly accumulate, altering cell behavior and undermining experimental reproducibility—often without obvious warning. In particular, human pluripotent stem cells, including iPSC and ESC, are prone to culture-acquired chromosomal gains and losses.
Researchers therefore face a critical question: how can every chromosomal alteration be monitored efficiently and accurately before it impacts outcomes?
Why Copy Number Changes Matter
History shows that copy number changes are not merely technical findings—they can directly shape biological outcomes. In 1959, the discovery that an extra copy of chromosome 21 causes Down syndrome demonstrated the impact of gains. In stem cells, alterations on chromosomes 12, 17, 20, or X can disrupt pluripotency or skew differentiation, while in cancer, amplifications such as HER2 or deletions like TP53 can drive disease progression.
Even low-level mosaicism can influence experimental outcomes. Understanding where and how these changes arise is essential for interpreting results and ensuring studies are built on a stable, well-characterized genomic foundation.
MLPA: When the Genome Learned to Speak in Numbers
For a long time, the genome spoke in pictures. Cytogeneticists learned to read its shapes under a microscope—stretched chromosomes, broken arms, missing pieces. Later, FISH lit up specific regions like stars in the night sky. Powerful, yes—but still selective. The genome was visible, but only where we chose to look.
MLPA reframed the question. Instead of asking the genome “Do you look normal?” MLPA asked a quieter, more precise question: “How many copies are really there?”
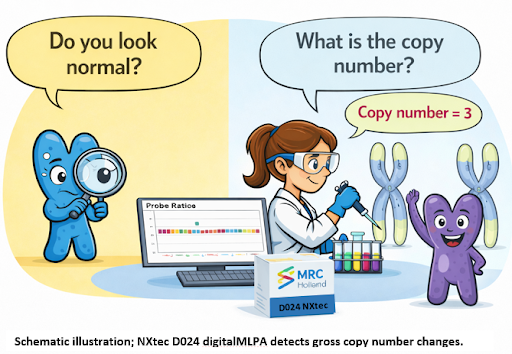
MLPA turned DNA copy numbers into measurable signals, introducing a quantitative molecular language. Each probe pair ligates only when its target is present, and signals rise or fall proportionally to genomic gain or loss—without requiring metaphase spreads or dividing cells. This made subtle deletions and duplications, previously invisible to karyotyping or routine FISH, clearly detectable. With the evolution to digitalMLPA, the same chemistry can now monitor hundreds to over thousands of targets, enabling sensitive genome-wide surveillance. D024 builds on this, detecting low-level mosaic copy number changes, providing early warning of genomic drift in long-term cultures, stem cell models (iPSC and ESC), and other sensitive research systems.
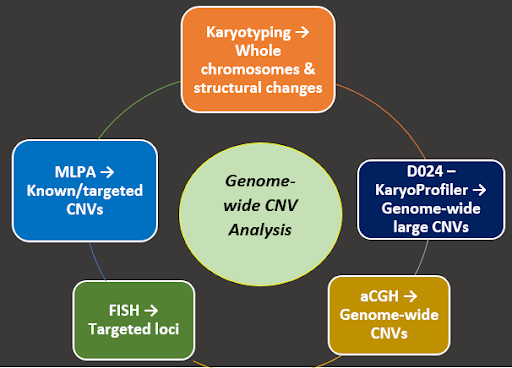
D024 KaryoProfiler: The Genome’s Detective
The D024 KaryoProfiler by MRC Holland is a digitalMLPA-based, genome-wide copy number assay designed to detect chromosomal gains and losses across most chromosome arms. By combining digitalMLPA chemistry with NGS readout on Next Generation Sequencing platforms, D024 enables scalable, genome-wide copy number profiling without the need for dividing cells, bridging classical cytogenetics and modern molecular analysis.
In a benchmark study using 43 cell line–derived samples from the Coriell CNVPANEL01, the NXtec D024 KaryoProfiler achieved 98.5% concordance with known cytogenetic profiles, supporting its reliability for genome-wide copy number detection.
When the genome whispers its secrets, scientists need a keen listener. The NXtec D024 KaryoProfiler acts as a watchful sentinel, scanning chromosomes to detect copy number changes as small as ~2–4 Mb—even in non-dividing cells.

D024 covers most chromosome arms with 1,180 genome-wide probes, plus additional control and reference probes for quality checks and cross-contamination detection. Whole chromosomes and large subchromosomal regions are monitored rapidly, providing a genome-wide snapshot in just 2–3 days—helping researchers track genomic stability in stem cell studies, disease models, and research aimed at developing new therapies.
Key note: D024 KaryoProfiler detects copy number changes (gains and losses) only. It does not detect balanced chromosomal rearrangements such as translocations or inversions, which require complementary methods like classical karyotyping or targeted FISH.
With this probe set, D024 identifies subtle genomic changes that could affect pluripotency, skew differentiation, or impact reproducibility. Its workflow integrates seamlessly into modern genomics labs, providing reliable genome-wide insight through automated analysis with Coffalyser digitalMLPA™ software—no advanced bioinformatics expertise required. https://www.dssimage.com/product/nxtec-d024-karyoprofiler/
Key Features of D024
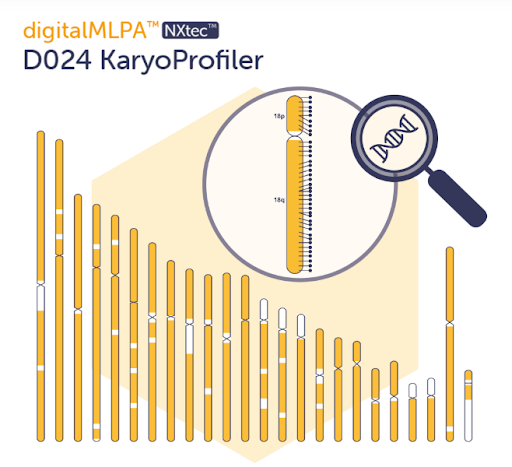
- Detects gains and losses across most chromosome arms (~2–4 Mb resolution)
- Works in non-dividing cells—no metaphase preparation required
- Results in approximately 2–3 days, depending on workflow
- Complements karyotyping, FISH, microarrays and NGS
- Automated analysis via Coffalyser digitalMLPA™
D024 KaryoProfiler is particularly valuable for:
- Monitoring genomic stability in long-term cell cultures
- Human pluripotent stem cell research (iPSC and ESC)
- Detection of culture-acquired chromosomal gains and losses
- Follow-up of abnormal karyotyping, FISH, or array CGH findings
- Research workflows requiring genome-wide copy number screening
Complementary MLPA Panels
While D024 provides a broad genome-wide overview, targeted digitalMLPA™ panels add precision:
| Panel | Focus | Highlight |
|---|---|---|
| D007 (ALL) | Acute lymphoblastic leukemia | Detects gene- and region-specific changes including hyperdiploidy and hypodiploidy |
| D006 (Multiple Myeloma) | Recurrent CNAs in Multiple Myeloma | Monitors clinically relevant alterations |
| P095 | Chromosomes 13, 18, 21, X, Y | Rapid aneuploidy screening |
Together, D024 and these panels provide a complete genomic toolkit—broad coverage, precise detection, and reliable insight for every experiment.
Fast, Reliable, High-Throughput
D024’s combination of speed, resolution, and easy workflow makes it ideal for routine genomic surveillance, follow-up of abnormal FISH, array CGH, or NGS results, and monitoring genomic drift in long-term cultures. With this approach, laboratories gain clarity, stability, and actionable insight into their samples.
Integrating D024 into DSS’s Complete Cytogenetics Workflow

Within DSS Imagetech’s integrated cytogenetics and genomics portfolio, D024 KaryoProfiler® serves as the genome-wide copy number backbone, complementing chromosome visualization, targeted FISH analysis, and advanced digital imaging systems.
Combined with DSS’s advanced microscopy from Evident and digital analysis software from Applied Spectral Imaging, laboratories can efficiently visualize chromosomes and streamline the detection of structural abnormalities.
For targeted FISH confirmation, DSS provides Abbott’s Vysis probes, including the AneuVysion kit for prenatal chromosome screening, along with ThermoBrite hybridization systems and VIP 2000 slide-processing systems and NeoDx CyFISH probes. Also media and reagents from EuroClone, further expand the FISH portfolio, supported by hybridization platforms such as HyperChrome (EuroClone). Automation solutions from ADS Biotec enhance efficiency and standardization across cytogenetics workflows.
Together, these solutions form a complete, integrated cytogenetics workflow, offering comprehensive chromosomal insights with faster turnaround and reproducible results.
Experience smarter, faster, and more precise cytogenetics and molecular genome analysis with DSS Imagetech: www.dssimage.com.
Closing Thoughts
From classical karyotyping to targeted molecular cytogenetic methods such as FISH, and copy number-focused molecular techniques like MLPA and microarrays, the study of chromosomes has advanced tremendously. The NXtec D024 KaryoProfiler now provides a robust, genome-wide assessment of copy number changes, including subtle gains, losses, and mosaic events. Complementing classical karyotyping and other cytogenetic approaches, D024 enables laboratories to confidently detect genomic alterations, clarify unclear karyotypes, and monitor genomic stability, giving researchers a clearer, actionable view of the genome.
In the quiet hours of the lab, the genome never stops working—and with D024, scientists can keep pace. Every experiment, culture, and study rests on a foundation of clarity, stability, and reliable insight.
Because what you can’t see can still change everything.
For more information on the NXtec D024 KaryoProfiler and its integration into your laboratory workflow, please contact DSS Imagetech at enquiry@dssimage.com.
References
- Tjio JH, Levan A. The chromosome number of man. Hereditas. 1956;42(1):1–6.
DOI: https://doi.org/10.1016/0002-9378(78)90337-x - Jacobs PA. Chromosome abnormalities and congenital disorders. Nat Rev Genet. 2010;11:87–100.
PMC: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4373004/ - Scarr S. Fluorescence in situ hybridization (FISH) – Chromosome painting. Memorial University.
https://www.mun.ca/biology/scarr/FISH_chromosome_painting.html - Vitillo L, et al. Multiplex ligation-dependent probe amplification (MLPA) for genome-wide copy number analysis. Methods Mol Biol. 2012;838:129–144.
PMC: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7122835/ - Kallioniemi A, et al. Comparative genomic hybridization for genome-wide DNA copy number analysis. Science. 1992;258(5083):818–821.
PMC: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1016317/ - Genomics Knowledge Hub. Microarray (array CGH) overview.
https://genomics.nshcs.org.uk/genotes/knowledge-hub/microarray-array-cgh/?utm_source=chatgpt.com - Schena M, et al. Microarray technology in molecular cytogenetics. Nat Genet. 2003;33 Suppl:1–5.
PMC: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2276893/?utm_source=chatgpt.com - Vitillo L, et al. Genomic stability in stem cells. Nat Rev Mol Cell Biol. 2020;21:413–426.
https://www.nature.com/articles/s41580-020-00292-z?utm_source=chatgpt.com - History of Down Syndrome Research. Trisomy 21 Research Society.
https://www.t21rs.org/history-of-down-syndrome-research/?utm_source=chatgpt.com - Vitillo L, et al. Copy number alterations in cancer and stem cells. Front Cell Dev Biol. 2019;7:57.
PMC: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6900480/ - MRC Holland. digitalMLPA™ technology overview.
https://www.mrcholland.com/technology/digitalmlpa?utm_source=chatgpt.com - DSS Imagetech. Integrated cytogenetics and genomics solutions.
https://www.dssimage.com/
About the Writer

Ms. Megha Dhumal is a Product Specialist – Clinical Diagnostics (Molecular), with 8+ years of experience in molecular diagnostics. She specializes in hematologic malignancy testing using real-time PCR, capillary electrophoresis, and NGS, with expertise spanning across solid tumors, genetic disorders, and infectious diseases. In her current role, she supports laboratories in implementing and optimizing integrated molecular diagnostic workflows, enabling accurate and reliable molecular results.
Latest Articles
Beyond the Microscope: Detecting Genomic Changes with D024 KaryoProfiler
BY DSS Imagetech Pvt Ltd February 17, 2026
In the Quiet Hours of the Lab In the quiet hours of a cytogenetics laboratory, cells are busy at work while no one is watching. They divide, adapt—and sometimes, silently,...
Read MoreWorld Cancer Day (4 February): Together, We Can Beat Cancer
BY DSS Imagetech Pvt Ltd February 4, 2026
Standing Together Against Cancer Cancer continues to affect millions of lives worldwide, touching not only patients but also families, caregivers, and entire communities. Despite the continued immense burden of cancer,...
Read MoreThe Unsung Hero of the Bench: Why the Evident CX23...
BY DSS Imagetech Pvt Ltd January 19, 2026
You know that feeling when you walk into a laboratory first thing in the morning? The hum of the refrigerator, the smell of ethanol and agar, the quiet potential of...
Read More